Karaca Lab. on Computational Structural Biology
Experimental structure determination techniques have solved tens of thousands of structures of biomolecules and their complexes. Still, the field cannot keep pace with the speed at which data are generated in other disciplines, such as genetics, biochemistry, and various associated “omics” technologies. Computational Structural Biology emerged to help overcome this bottleneck to generate high-accuracy structural models rapidly.
Since late 2020, we have witnessed a revolution in structure prediction with the emergence of AlphaFold. Together with AF and other AI-based tools, we are closer to interpreting the omics data at the structural level than ever. In our lab, we are developing computational tools and approaches to assist this interpretation by focusing on the structure-function relationships of biomolecular complexes.
Research Interests
We are particularly interested in understanding the role of dynamics and variations across protein interfaces since we think that such an understanding will pave the way to elucidate the ruleset of biomolecular recognition.
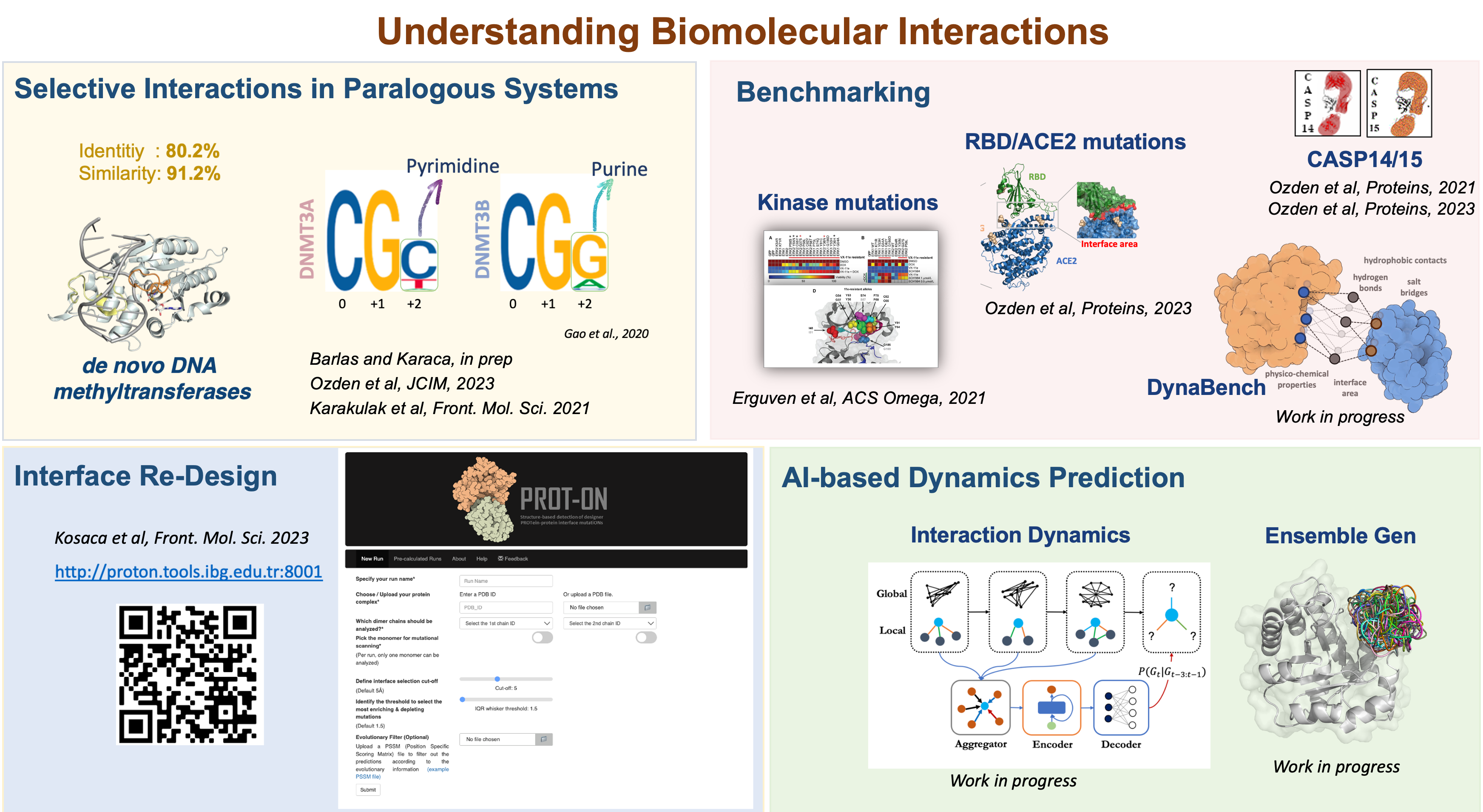
We perform our research under four themes:
- Concentrating on interface dynamics to unravel selective interactions leading to paralogous complex formation, such as in selective transcription factor interactions or specific interactions happening in signaling cascades.
- Benchmarking (i) the accuracy of mutation predictions for clinically relevant proteins, as done for kinases and SARS-CoV-2. (ii) the accuracy of protein complex prediction tools, as done for the CASP14 and CASP15 multimeric assessment categories.
- Finding optimal mutations over existing interfaces for designing new protein binders. The recent PROT-ON tool we developed for this can be reached through our webserver.
- Developing AI-based tools to predict interface dynamics and conformational landscape of proteins in an economical manner (work in progress).
For more, you check our Github Page, Dr. Karaca's Twitter account, or her Google Scholar Profile.
Brief CV of the PI
Education:
- Ph.D. in Computational Structural Biology, Utrecht University (Bonvin Lab, the Netherlands)
- M.Sc. & B.Sc. in Chemical Engineering, Bogazici University (Haliloglu Lab, Türkiye)
Experience:
- Since 2017 Asst. Prof. in Dokuz Eylul University & PI in Izmir Biomedicine and Genome Center
- 2013-2016 EMBL Heidelberg (Carlomagno & Barabas Labs, Germany)
- 2006-2007, Summer terms, NCI (USA, Nussinov Lab)
Link to the full CV of Dr. Karaca (last update was made on 2023-12).
OUTREACH ACTIVITIES
Selected presentations:
- Nature's news piece on the CASP15 conference held in Antalya (2022).
- CASP15 assembly assessment presentation (2022).
- EBI webinar on the impact of AlphaFold in training in life sciences, where Dr. Karaca was a panelist (2022).
- JOBIM 2021 - Symposium of Integrative Structural Modeling, where Dr. Karaca presented their work on how nucleosome dynamics selects its transcription factor binder Sox.
- The assembly assessment presentation of CASP14 can be downloaded from here (late 2020)
- Which factors lie behind AlphaFold's success, presented on the Youtube Channel of the Science Academy of Turkey (in Turkish, Late 2020)
- Exploring the molecular machines, presented on the International Day of Women and Girls in Science on the Youtube Channel of the Scientific and Technological Research Council of Turkey (in Turkish, Early 2021)
- Dr. Karaca's presentation on her postdoctoral work presented in ISCB RSG Turkey's webinar series (2018)
Group Members

Karaca Lab. on Computational Structural Biology
Research Group Leader
Ezgi KARACA
ezgi.karaca@ibg.edu.tr
+90 232 299 41 00
(5041)
+9
0 (232) 299 _ _

Burcu ÖZDEN
Research Assistant
burcu.ozden@ibg.edu.tr

Büşra SAVAŞ
PhD Student
busra.savas@ibg.edu.tr

Buse ŞAHİN
MSc Student
buse.sahin@ibg.edu.tr

Murat Taylan ŞAHİN
MSc Student
taylan.sahin@std.ibg.edu.tr

Gülce Berfin ERCAN
Undergraduate Student
None

Ayşe Berçin BARLAS YILMAZ
Post-Doc Researcher
aysebercin.barlas@ibg.edu.tr

İbrahim Atakan KUBİLAY
PhD Student
atakan.kubilay@std.ibg.edu.tr
Former Members

Mehmet ERGÜVEN
Research Assistant
mehmet.erguven@msfr.ibg.edu.tr

Eda ŞAMİLOĞLU TENGİRŞEK
Research Assistant
eda.samiloglu@ibg.edu.tr

Beyza KAYNARCA
Research Assistant
beyza.kaynarca@ibg.edu.tr

Tülay KARAKULAK
Researcher
tulay.karakulak@ibg.edu.tr

Ayşe Berçin BARLAS YILMAZ
PhD Student
aysebercin.barlas@ibg.edu.tr

Deniz DOĞAN
MSc Student
deniz.dogan@msfr.ibg.edu.tr

Can YÜKRÜK
Undergraduate Student
can.yukruk@msfr.ibg.edu.tr

Can YÜKRÜK
Visiting Student
can.yukruk@msfr.ibg.edu.tr

İrem YILMAZBİLEK
Undergraduate Student
irem.yilmazbilek@ibg.edu.tr

Barış Can EKİM
PhD Student
bariscan.ekim@msfr.ibg.edu.tr
+90 232 299 41
00
(0 (539) 384 29 26)

Melis OKTAYOĞLU
Undergraduate Student
melis.oktayoglu@ibg.edu.tr

Aleyna Dilan KIRAN
Undergraduate Student
aleyna.kiran@msfr.ibg.edu.tr

Melis OKTAYOĞLU
Undergraduate Student
melis.oktayoglu@ibg.edu.tr

Aybike ERDOĞAN
PhD Student
aybike.erdogan@ibg.edu.tr

İrem YILMAZBİLEK
MSc Student
irem.yilmazbilek@ibg.edu.tr

Atakan ÖZSAN
Undergraduate Student
atakan.ozsan@ibg.edu.tr

Beyza KAYNARCA
Undergraduate Student
beyza.kaynarca@ibg.edu.tr

Mehdi KOŞACA
PhD Student
mehdi.kosaca@ibg.edu.tr

Beyza KAYNARCA
Visiting Researcher
beyza.kaynarca@ibg.edu.tr

Hilal TAY
MSc Student
hilal.tay@ibg.edu.tr

Atakan ÖZSAN
Undergraduate Student
atakan.ozsan@ibg.edu.tr

Tuğçe İNAN
Post-Doc Researcher
tugce.inan@ibg.edu.tr

İrem DEMİR
Undergraduate Student
irem.demir@std.ibg.edu.tr

Mennan GÖK
Undergraduate Student
mennan.gok@std.ibg.edu.tr
Selected Publications
Koşaca M, Yılmazbilek İ, Karaca E. PROT-ON: A structure-based detection of designer PROTein interface MutatiONs.. Frontiers in molecular biosciences. 2023 March ; 10 : 1063971. doi:10.3389/fmolb.2023.1063971. Download
Ozden B, Boopathi R, Barlas AB, Lone IN, Bednar J, Petosa C, Kale S, Hamiche A, Angelov D, Dimitrov S, Karaca E. Molecular Mechanism of Nucleosome Recognition by the Pioneer Transcription Factor Sox.. Journal of chemical information and modeling. 2023 June ; 63 (12) : 3839-3853. doi:10.1021/acs.jcim.2c01520. Download
Ozden B, Şamiloğlu E, Özsan A, Erguven M, Yükrük C, Koşaca M, Oktayoğlu M, Menteş M, Arslan N, Karakülah G, Barlas AB, Savaş B, Karaca E. Benchmarking the accuracy of structure-based binding affinity predictors on Spike-ACE2 deep mutational interaction set.. Proteins. 2023 November . doi:10.1002/prot.26645. Download
Erguven M, Kilic S, Karaca E, Diril MK.. Genetic complementation screening and molecular docking give new insight on phosphorylation-dependent Mastl kinase activation. Journal of Biomolecular Structure and Dynamics. 2022 October . doi:10.1080/07391102.2022.2131627. Download
Karaca E, Prévost C, Sacquin-Mora S. Modeling the Dynamics of Protein-Protein Interfaces, How and Why?. Molecules (Basel, Switzerland). 2022 March ; 27 (6) : 1841. doi:10.3390/molecules27061841. Download
Erguven M, Cornelissen NV, Peters A, Karaca E, Rentmeister A. Enzymatic Generation of Double-Modified AdoMet Analogues and Their Application in Cascade Reactions with Different Methyltransferases.. Chembiochem : a European journal of chemical biology. 2022 December ; 23 (24) : e202200511. doi:10.1002/cbic.202200511. Download
Cingöz S, Soydemir D, Öner TÖ, Karaca E, Özden B, Kurul SH, Bayram E, University of Washington Center for Mendelian Genomics, Coe BP, Nickerson DA, Eichler EE. Novel biallelic variants affecting the OTU domain of the gene OTUD6B associate with severe intellectual disability syndrome and molecular dynamics simulations.. European journal of medical genetics. 2022 June ; 65 (6) : 104497. doi:10.1016/j.ejmg.2022.104497. Download
HIZ, AYŞE SEMRA; KILIÇ, SEVAL; BADEMCİ, GÜNEY; KARAKULAK, TÜLAY; ERDOĞAN, AYBİKE; YÜCEL, BURCU ÖZDEN; YAZICIOĞLU, ÇİĞDEM ERESEN; BAĞRIYANIK, ŞERİFE ESRA ERDAL; YİŞ, ULUÇ; TEKİN, MUSTAFA; KARAKÜLAH, GÖKHAN; EREK, EZGİ KARACA; and ÖZTÜRK, MEHMET. VARS1 Mutations Associated with Neurodevelopmental Disorder Are Located on A Short Amino Acid Stretch of The Anticodon-Binding Domain. Turkish Journal of Biology. 2022 January ; 46 (6) : 458-464. doi:10.55730/1300-0152.2631. Download
Erguven M, Karakulak T, Diril MK, Karaca E. How Far Are We from the Rapid Prediction of Drug Resistance Arising Due to Kinase Mutations?. ACS omega. 2021 January ; 6 (2) : 1254-1265. doi:10.1021/acsomega.0c04672. Download
Karakulak T, Rifaioglu AS, Rodrigues JPGLM, Karaca E. Predicting the Specificity- Determining Positions of Receptor Tyrosine Kinase Axl. Frontiers in Molecular Biosciences. 2021 June (8:658906) . doi:10.3389/fmolb.2021.658906. Download
Serdal Gungor, Yavuz Oktay, Semra Hiz, Álvaro Aranguren-Ibáñez, Ipek Kalafatcilar, Ahmet Yaramis, Ezgi Karaca, Uluc Yis, Ece Sonmezler, Burcu Ekinci, Mahmut Aslan, Elmasnur Yilmaz, Sunitha Balaraju, Nora Szabo, Steven Laurie, Sergi Beltran, Daniel G. MacArthur, Denisa Hathazi, Rita Horvath. Autosomal recessive variants in TUBGCP2 alter the γ-tubulin ring complex leading to neurodevelopmental disease. iScience. 2021 January ; 24 (1) . doi:10.1016/j.isci.2020.101948. Download
Ozden B, Kryshtafovych A, Karaca E. Assessment of the CASP14 assembly predictions.. Proteins. 2021 August . doi:10.1002/prot.26199. Download
Circir A, Koksal Bicakci G, Savas B, Doken DN, Henden ŞO, Can T, Karaca E, Erson-Bensan AE. A C-term truncated EIF2Bγ protein encoded by an intronically polyadenylated isoform introduces unfavorable EIF2Bγ-EIF2γ interactions.. Proteins. 2021 November . doi:10.1002/prot.26284. Download
Azbazdar Y, Ozalp O, Sezgin E, Veerapathiran S, Duncan AL, Sansom MSP, Eggeling C, Wohland T, Karaca E, Ozhan G. More Favorable Palmitic Acid Over Palmitoleic Acid Modification of Wnt3 Ensures Its Localization and Activity in Plasma Membrane Domains. Frontiers in cell and developmental biology. 2019 November ; 7 : 281. doi:10.3389/fcell.2019.00281. Download
Pavlopoulou A, Karaca E, Balestrazzi A, Georgakilas AG. In Silico Phylogenetic and Structural Analyses of Plant Endogenous Danger Signaling Molecules upon Stress. Oxidative medicine and cellular longevity. 2019 July ; 2019 : 8683054. doi:10.1155/2019/8683054. Download
Dafsari HS, Sprute R, Wunderlich G, Daimagüler HS, Karaca E, Contreras A, Becker K, Schulze-Rhonhof M, Kiening K, Karakulak T, Kloss M, Horn A, Pauls A, Nürnberg P, Altmüller J, Thiele H, Assmann B, Koy A, Cirak S. Novel mutations in KMT2B offer pathophysiological insights into childhood-onset progressive dystonia.. Journal of human genetics. 2019 August ; 64 (8) : 803-813. doi:10.1038/s10038-019-0625-1. Download
Bonvin AMJJ, Karaca E, Kastritis PL, Rodrigues JPGLM. Defining distance restraints in HADDOCK. Nature Protocols. 2018 June ; 13 : 1503. doi:10.1038/s41596-018-0017-6. Download
Rubio-Cosials A, Schulz EC, Lambertsen L, Smyshlyaev G, Rojas-Cordova C, Forslund K, Karaca E, Bebel A, Bork P, Barabas O. Transposase-DNA Complex Structures Reveal Mechanisms for Conjugative Transposition of Antibiotic Resistance. Cell. 2018 March ; 173 (1) : 208-220.e220. doi:10.1016/j.cell.2018.02.032. Download
Ercan I, Tufekci KU, Karaca E, Genc S, Genc K. Peptide Derivatives of Erythropoietin in the Treatment of Neuroinflammation and Neurodegeneration. Advances in Protein Chemistry and Structural Biology. 2018 February ; 112 : 309-357. doi:10.1016/bs.apcsb.2018.01.007. Download
Athanasia Pavlopoulou, Ezgi Karaca, Alma Balestrazzi, Alexandros G. Georgakilas. InSilicoPhylogenetic andStructural Analysis ofPlant Damps. Preprints. 2018 July ; 2018070522 : 1-14. doi:10.20944/preprints201807.0522.v1. Download
Karaca E, Rodrigues JPGLM, Graziadei A, Bonvin AMJJ, Carlomagno T. M3: an integrative framework for structure determination of molecular machines. Nature methods. 2017 August ; 14 (9) : 897-902. doi:10.1038/nmeth.4392. Download
Vangone A, Rodrigues JP, Xue LC, van Zundert GC, Geng C, Kurkcuoglu Z, Nellen M, Narasimhan S, Karaca E, van Dijk M, Melquiond AS, Visscher KM, Trellet M, Kastritis PL, Bonvin AM. Sense and simplicity in HADDOCK scoring: Lessons from CASP-CAPRI round 1.. Proteins. 2017 March ; 85 (3) : 417-423. doi:10.1002/prot.25198. Download
Bebel A, Karaca E, Kumar B, Stark WM, Barabas O. Structural snapshots of Xer recombination reveal activation by synaptic complex remodeling and DNA bending.. eLife. 2016 December ; 5 . doi:10.7554/eLife.19706. Download
van Zundert GCP, Rodrigues JPGLM, Trellet M, Schmitz C, Kastritis PL, Karaca E, Melquiond ASJ, van Dijk M, de Vries SJ, Bonvin AMJJ. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes.. Journal of molecular biology. 2016 February ; 428 (4) : 720-725. doi:10.1016/j.jmb.2015.09.014. Download
Rodrigues JP, Karaca E, Bonvin AM. Information-driven structural modelling of protein-protein interactions.. Methods in molecular biology (Clifton, N.J.). 2015 January ; 1215 : 399-424. doi:10.1007/978-1-4939-1465-4_18. Download
Lensink MF, Moal IH, Bates PA, Kastritis PL, Melquiond AS, Karaca E, Schmitz C, van Dijk M, Bonvin AM, Eisenstein M, Jiménez-García B, Grosdidier S, Solernou A, Pérez-Cano L, Pallara C, Fernández-Recio J, Xu J, Muthu P, Praneeth Kilambi K, Gray JJ, Grudinin S, Derevyanko G, Mitchell JC, Wieting J, Kanamori E, Tsuchiya Y, Murakami Y, Sarmiento J, Standley DM, Shirota M, Kinoshita K, Nakamura H, Chavent M, Ritchie DW, Park H, Ko J, Lee H, Seok C, Shen Y, Kozakov D, Vajda S, Kundrotas PJ, Vakser IA, Pierce BG, Hwang H, Vreven T, Weng Z, Buch I, Farkash E, Wolfson HJ, Zacharias M, Qin S, Zhou HX, Huang SY, Zou X, Wojdyla JA, Kleanthous C, Wodak SJ. Blind prediction of interfacial water positions in CAPRI.. Proteins. 2014 April ; 82 (4) : 620-32. doi:10.1002/prot.24439. Download
Lensink MF, Brysbaert G, Nadzirin N, Velankar S, Chaleil RAG, Gerguri T, Bates PA, Laine E, Carbone A, Grudinin S, Kong R, Liu RR, Xu XM, Shi H, Chang S, Eisenstein M, Karczynska A, Czaplewski C, Lubecka E, Lipska A, Krupa P, Mozolewska M, Golon Ł, Samsonov S, Liwo A, Crivelli S, Pagès G, Karasikov M, Kadukova M, Yan Y, Huang SY, Rosell M, Rodríguez-Lumbreras LA, Romero-Durana M, Díaz-Bueno L, Fernandez-Recio J, Christoffer C, Terashi G, Shin WH, Aderinwale T, Maddhuri Venkata Subraman SR, Kihara D, Kozakov D, Vajda S, Porter K, Padhorny D, Desta I, Beglov D, Ignatov M, Kotelnikov S, Moal IH, Ritchie DW, Chauvot de Beauchêne I, Maigret B, Devignes MD, Ruiz Echartea ME, Barradas-Bautista D, Cao Z, Cavallo L, Oliva R, Cao Y, Shen Y, Baek M, Park T, Woo H, Seok C, Braitbard M, Bitton L, Scheidman-Duhovny D, Dapkūnas J, Olechnovič K, Venclovas Č, Kundrotas PJ, Belkin S, Chakravarty D, Badal VD, Vakser IA, Vreven T, Vangaveti S, Borrman T, Weng Z, Guest JD, Gowthaman R, Pierce BG, Xu X, Duan R, Qiu L, Hou J, Ryan Merideth B, Ma Z, Cheng J, Zou X, Koukos PI, Roel-Touris J, Ambrosetti F, Geng C, Schaarschmidt J, Trellet ME, Melquiond ASJ, Xue L, Jiménez-García B, van Noort CW, Honorato RV, Bonvin AMJJ, Wodak SJ. Blind prediction of homo- and hetero-protein complexes: The CASP13-CAPRI experiment.. Proteins. 2014 December ; 87 (12) : 1200-1221. doi:10.1002/prot.25838. Download
Rodrigues JP, Melquiond AS, Karaca E, Trellet M, van Dijk M, van Zundert GC, Schmitz C, de Vries SJ, Bordogna A, Bonati L, Kastritis PL, Bonvin AM. Defining the limits of homology modeling in information-driven protein docking.. Proteins. 2013 December ; 81 (12) : 2119-28. doi:10.1002/prot.24382. Download
Karaca E, Bonvin AM. On the usefulness of ion-mobility mass spectrometry and SAXS data in scoring docking decoys.. Acta crystallographica. Section D, Biological crystallography. 2013 May ; 69 (Pt 5) : 683-94. doi:10.1107/S0907444913007063. Download
Karaca E, Bonvin AM. Advances in integrative modeling of biomolecular complexes.. Methods. 2013 March ; 59 (3) : 372-81. doi:10.1016/j.ymeth.2012.12.004. Download
Rodrigues JP, Trellet M, Schmitz C, Kastritis P, Karaca E, Melquiond AS, Bonvin AM. Clustering biomolecular complexes by residue contacts similarity.. Proteins. 2012 July ; 80 (7) : 1810-7. doi:10.1002/prot.24078. Download
Karaca E, Bonvin AM. A multidomain flexible docking approach to deal with large conformational changes in the modeling of biomolecular complexes.. Structure. 2011 April ; 19 (4) : 555-65. doi:10.1016/j.str.2011.01.014. Download
Karaca E, Tozluoğlu M, Nussinov R, Haliloğlu T. Alternative allosteric mechanisms can regulate the substrate and E2 in SUMO conjugation.. Journal of molecular biology. 2011 March ; 406 (4) : 620-30. doi:10.1016/j.jmb.2010.12.044. Download
Adrien S.J. Melquiond, Ezgi Karaca, Panagiotis L. Kastritis, Alexandre M.J.J. Bonvin. Next challenges in protein–protein docking: from proteome to interactome and beyond. WIREs Computational Molecular Science. 2011 January . doi:10.1002/wcms.91. Download
de Vries SJ, Melquiond AS, Kastritis PL, Karaca E, Bordogna A, van Dijk M, Rodrigues JP, Bonvin AM. Strengths and weaknesses of data-driven docking in critical assessment of prediction of interactions.. Proteins. 2010 November ; 78 (15) : 3242-9. doi:10.1002/prot.22814. Download
Karaca E, Melquiond AS, de Vries SJ, Kastritis PL, Bonvin AM. Building macromolecular assemblies by information-driven docking: introducing the HADDOCK multibody docking server.. Molecular and Cellular proteomics. 2010 August ; 9 (8) : 1784-94. doi:10.1074/mcp.M000051-MCP201. Download
Nicastro G, Todi SV, Karaca E, Bonvin AM, Paulson HL, Pastore A. Understanding the role of the Josephin domain in the PolyUb binding and cleavage properties of ataxin-3.. PloS one. 2010 August ; 5 (8) : e12430. doi:10.1371/journal.pone.0012430. Download
Tozluoğlu M, Karaca E, Nussinov R, Haliloğlu T. A mechanistic view of the role of E3 in sumoylation.. PLoS computational biology. 2010 August ; 6 (8) . doi:10.1371/journal.pcbi.1000913. Download
Tozluoğlu M, Karaca E, Haliloglu T, Nussinov R. Cataloging and organizing p73 interactions in cell cycle arrest and apoptosis.. Nucleic acids research. 2008 September ; 36 (15) : 5033-49. doi:10.1093/nar/gkn481. Download
Total : 39
Projects
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : Kornea Endoteli Dokusunun Kök Hücre ve Biyomühendislik Yaklasımlarıyla Geliştirilmesi, Ongoing
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : Interaction of Spesific Insulin Receptor Isoforms with c-Met: Mechanisms and Contributing Roles in the Progression of Hepatocellular Carcinoma, Ongoing
Health Institutes of Turkey- TUSEB - RD : PROT-ON: Structure-based Detection of Critical Mutations in Redesigning Protein-protein Interfaces, Ongoing
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : Epigenetics of Epithelial-to-Mesenchymal and Mesenchymal-to-Epithelial Transitions : Role of Histone Variants and Pioneer Transcription Factors, Ongoing
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : Systematic Approach to Characterize Pioneer Transcription Factor Binding to Chromatin, Finished
European Molecular Biology Organization - EMBO - RD : Identification of Pioneer Transcription Factor Binding Modes to Chromatin, Ongoing
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : İnsan DNA Metiltransferaz 3A Enziminin Çalışma İlkelerinin Hesaplamalı Yöntemlerle Tanımlanması, Finished
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : Development of Recombinant Protein Based Vaccine Against SARS-CoV-2, Ongoing
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : Development of Drugs Based on “Ghost Receptor-Antibody” Against SARS-CoV-2, Ongoing
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : TROPIC: An integrative approach to define druggable human-parasite interactions, Ongoing
Open Positions
POST-DOCTORAL RESEARCHER
We are looking for a post-doc researcher, who is expected to work on molecular modeling and/or method development. If you would like to be considered for the position, here are the main requirements.
- Extensive experience in molecular modeling OR in a scientific programming language (e.g. Python, MATLAB, R)
- Ability to work in an interdisciplinary environment.
Please contact ezgi.karaca@ibg.edu.tr and check Karaca Lab main page for further details.
SOFTWARE ENGINEER
We are looking for a full-time or part-time software engineer, who will be supporting the team in tool development, scripting and web-service design. If you would like to be considered for the position, here are the main requirements:
- Graduated from Computer Engineering, Software Engineering or Bioinformatics departments OR having an extensive experience in programming.
- Ability to work in an interdisciplinary environment.
Please contact ezgi.karaca@ibg.edu.tr and check Karaca Lab main page for further details.
Awards
- Installation Grant by EMBO, 2020
- Young Scientist (BAGEP) Award by Science Academy, 2020
- Young Investigator Travel Award by eLife Journal, 2019
Academic Memberships
- Tıp Bilişim Derneği, 2018
Contact

Karaca Lab. on Computational Structural Biology
Research Group Leader
Ezgi KARACA
ezgi.karaca@ibg.edu.tr
+90 232 299 41 00
(5041)
+9
0 (232) 299 _ _